+33 (0) 3 20 16 91 40
Studies from biology PTS

AGLAE processes data from series of proficiency tests and draws in particular information about the analytical methods. These studies aim to inform laboratories and help them understand the differences they may also observe. The following studies have been conducted in biology:
- Analysis of coliform bacteria and E.coli on CCA medium
- Precision models for basic microbiology and pathogens
- Legionella by PCR versus by culture
Detection and enumeration of coliform bacteria and E.coli - Uncertainty associated with the colour of the colonies observed on CCA medium
The international standard method to detect and enumerate in water coliform bacteria and more specifically Escherichia coli was modified at the end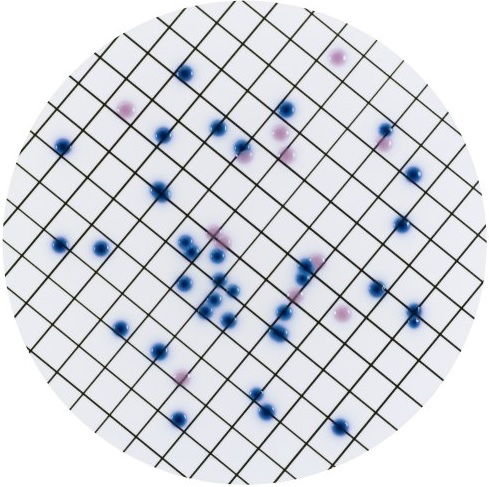 of 2014.
of 2014.
It consists from now on in using a type of agar medium new for the analysis of the environment: a chromogenic medium, Chromagar, for which the reading of colonies is based on the colonies colour. If the colony appears blue, it is Escherichia coli. If it is pink, it is another coliform bacteria.
What is the order of magnitude of the colour variations from one strain to another? And what is the uncertainty associated with the perception of this colour by the microbiologist?
In order to address these questions, and also evaluate the practicability and efficiency of this new method, AGLAE conducted experiments in 2015.
The results of this study based on a small number of strains are mixed.
- Positive point: despite of significant colour differences from one strain to another and despite of divergences between microbiologists in the perception of these colours, it seems that the pigmentation of the colonies centre is a reliable criterion.
- But it also appears that on small colonies the pigmentation is not sufficiently developed.
- Then, and this is a bit more disturbing, among the tested strains, several presented singular responses; in particular for the distinction of Escherichia coli among other coliform bacteria.
At the French level, this method is not enforced yet but it could be in the future; depending on what the public authorities in charge of the sanitary control will decide.
Download the study Analysis of coliform bacteria and E.coli on CCA medium - colour uncertainty (only abstract in English)
Precision models for basic microbiology, Ps. aeruginosa and pathogenic staphylococci in clean waters - Outcomes on the uncertainties
Almost 15 years of experience were processed to help you understand and interpret the precision models in microbiology and for some pathogens in clean waters.
The evolution of the reproducibility depending on the bacterial load level is presented in order to statistically comprehend the variability of bacterial enumeration during routine analysise. More precisely, the following paramters were concerned: coliform bacteria, Escherichia coli, culturable micro-organisms at 22°C and 36°C, intestinal enterococci, spores of sulfite-reducing anaerobes, Pseudomonas aeruginosa and pathogenic staphylococci.

The matrices used to prepare the test samples were chlorinated and neutralized drinking waters or only neutralized for some tests. For most parameters, spiking had been carried out with strains grown in broth.
The direct links with the uncertainty of measurement calculations at the profession scale and the perspectives in terms of calculation tools are also discussed at the end of the document.
Download the study Precision models for basic microbiology and pathogens (in English)
Detection and enumeration of Legionella spp and of Legionella pneumophila: quantitative PCR versus culture on GVPC medium, an example taken from AGLAE's proficiency testing
In 2013, AGLAE sent the same test material (hot domestic water) on the one hand to laboratories (263) carrying out the Legionella analysis by culture on GVPC medium, and on the other hand to laboratories (36) using PCR. The study of the results obtained enables us to compare the two methods in terms of response level and precision (repeatability but mainly reproducibility).
The conclusions of this example match the observations already made, in particular those published in scientific literature:
- A 10 factor has been observed between the measurand response ‘q-PCR’ and the measurand response ‘culture on GVPC’
- Regarding the reproducibility of one or the other methods, they were not found to be radically different. The reproducibility variation coefficient of the data in log is around 5% for both methods
This study confirms the interest to use q-PCR method as a quick detection and enumeration method for Legionella and Legionella pneumophila.
Download the study Legionella by PCR versus by culture (in French only)
